Working in the field has its limitations, as Alex Tolley reminds us in the essay that follows, but at least biologists have historically been on the same planet with their specimens. Today’s hottest news would be the discovery of life on another world, as we saw in the brief flurries over the Viking results in 1976 or the Martian meteorite ALH84001. We rely, of course, on remote testing and will increasingly count on computer routines that can make the fine distinctions needed to choose between biotic and abiotic reactions. A new technique recently put forward by Robert Hazen and James Cleaves holds great promise. Alex gives it a thorough examination including running tests of his own to point to the validity of the approach. One day using such methods on Mars or an ice giant moon may confirm that abiogenesis is not restricted to Earth, a finding that would have huge ramifications not just for our science but also our philosophy.
by Alex Tolley
Perseverance rover on Mars – composite image.
Cast your mind back to Darwin’s distant 5-year voyage on HMS Beagle. He could make very limited observations, make drawings and notes, and preserve his specimen collection for his return home to England.
Fifty years ago, a field biologist might not have much more to work with. Hours from a field station or lab with field guides and kits to preserve specimens, with no way to communicate. As for computers to make repetitive calculations, fuggedaboutit.
Fast forward to the late 20th and early 21st centuries, and fieldwork is extending out to the planets in our solar system to search for life. Like Darwin’s voyage, the missions are distant and long. Unlike Darwin, samples have not yet been returned from any planets, only asteroids and comets. Communication is slow, more on the order of field experiences. But instead of humans, our robot probes are “Going where no one has gone before” and humans may not go until much later. The greater the communication lag, the more problematic the central command to periphery control model. Reducing this delay demands a need for more peripheral autonomy at the periphery to make local decisions.
The 2006 Astrobiology Field Laboratory Science Steering Group report recommended that the Mars rover be a field laboratory, with more autonomy [17]. The current state of the art is the Perseverance rover taking samples in the Jezero crater, a prime site for possible biosignatures. Its biosignature instrument, SHERLOC, uses Raman spectrography and luminescence to detect and identify organic molecules [6]. While organic molecules may have been detected [19], the data had to be transmitted to Earth for interpretation, maintaining the problem of lag times between each sample to be chosen and analyzed.
As our technology improves, can these robots operating on planetary surfaces be able to do more effective in situ analyses in the search for extant or extinct life, so that they can operate more quickly like a human field scientist, in the search for life?
While we “know life when we see it”, nevertheless we still struggle to define what life is, although with terrestrial life we have sufficient characteristics except for edge cases like viruses and some ambiguous early fossil material. However, some defining characteristics do not apply to dead, or fossilized organisms and their traces. Fossil life does not metabolize, reproduce, or move, and molecules that are common to life no longer exist in their original form. Consider the “fossil microbes” on the Martian meteorite ALH84001 that caused such a sensation when announced but proved ambiguous.
Historically, for fossil life, we have relied on detecting biosignatures, such as C13/C12 ratios in minerals (due to chlorophyll carbon isotope preference), long-lasting biomolecules like lipids, homochirality of organic compounds, and disequilibria in atmospheric gases. Biomolecules can be ambiguous, as the amino acids detected in meteorites are most likely abiotic, something the Miller-Urey experiment demonstrated many decades ago.
Ideally, we would like a detection method that is simple, robust, and whose results can be interpreted locally without requiring analysis on Earth.
A new method to try to identify the probably biotic nature of samples with organic material is the subject of a new paper from a collaboration under Prof. Robert Hazen and James Cleaves. The team not only uses an analytical method—pyrolysis gas chromatography coupled to electron impact ionization mass spectrometry (Pyr-GC-EI-MS) to heat (pyrolyze), fractionate volatile components (gas chromatography), and determine their mass (mass spectrometry), but also analyzes the data to classify whether the new samples contain organic material of biological origin. Their reported early results are very encouraging [10, 11, 12].
The elegance of Hazen et al’s work has been to apply the Pyr-GC-EI-MS technique [3, 15, 18] that is not only available in the laboratory, but is also designed for planetary rovers to meet the need for local analysis. Their innovation has been to couple this process with computationally lightweight machine learning models to classify the samples, thereby bypassing the time lags associated with distant terrestrial interpretation. A rover could relatively rapidly take samples in an area and determine whether any might have a biosignature based on a suite of different detected compounds and make decisions locally on how to proceed.
The resulting data of masses and extraction time can be reduced and then classified using the pre-trained Random Forest [4], which is a suite of Decision Trees (see Figure 3) using samples of the feature set of masses, to provide a classification, which with the currently tested samples, provides a better than 90% probability of correct classification. The reported experiment used 134 samples, 75 labeled as abiotic and 59 as biotic or of biotic origin. The data set ranged in mass from 50 to 700 and several thousand scans over time. This data was reduced to a manageable size by reducing the mass and time ranges to 8147 values. The samples were then run against several machine learning methods, of which the Random Forest worked best.
To provide a visualization of which mass and time values were most instrumental in classifying the data, the 20 most informative data points were extracted and overlaid on the MS data as shown in Figure 1.
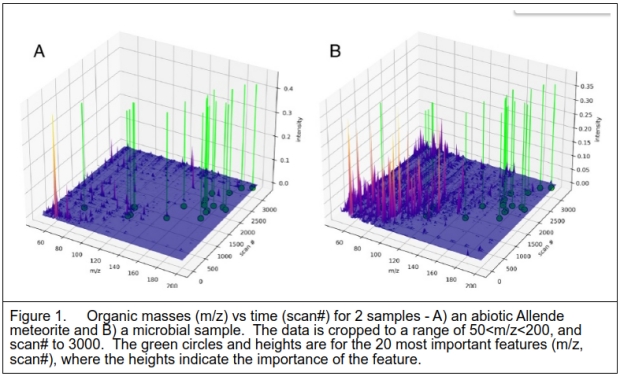
Figure 1 above shows 2 samples of data. One is the Allende meteorite which is classified as a 4.5 billion years old abiotic sample. This is contrasted with one of the microbial samples. While the details of the meteorite sample were not provided, older analyses by others indicated that the surface was contaminated with terrestrial material, whilst the interior matrix included polycyclic aromatic hydrocarbons, a common material found in samples from space missions [7,8]. The bacterial sample, as expected, shows many more compounds after pyrolysis, as the organism is composed of a large variety of organic compounds including amino acids, nucleobases, lipids, and sugars which will decompose with heating. A key point is that the discriminant features are not coincident with the most common masses in the samples, but rather in the rarer compounds as indicated by their intensities. [The lower bound mass bin ensures that common pyrolysis products such as low carbon number compounds will be excluded from the analysis and visualization. The data is normalized to the highest peak so that relative values rather than absolutes are analyzed to eliminate sample amounts.] Most of the defining compounds are in the 140 – 200 mass range, which would imply all-carbon compounds with 12-16 atoms.
Figure 2 shows a 2-dimensional Principal Components Analysis (PCA) using the 20 most informative features that illustrate the separation of the sample types. The expanded box encompasses all the abiotic samples.
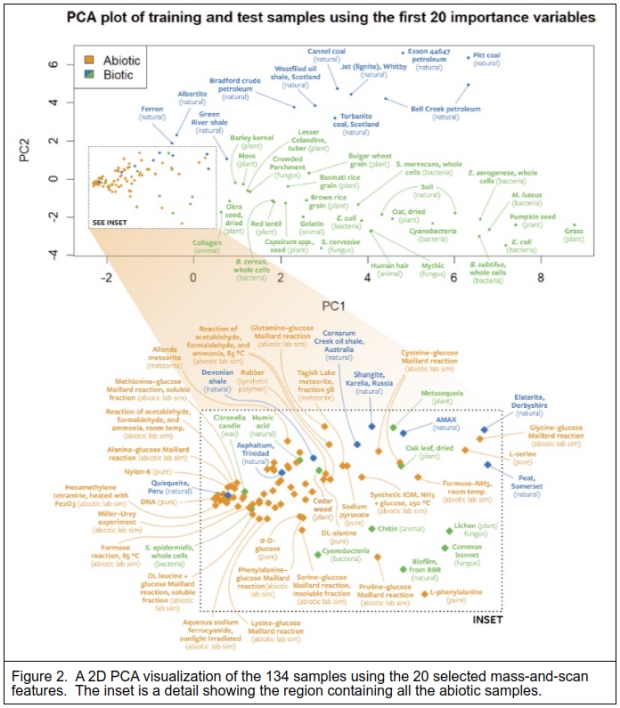
I note that even though the biotic and natural samples were given the same classification label, these samples separate quite clearly too, indicating that the natural samples appear almost distinct from the biotic samples. What is perhaps surprising is that biological materials like cedarwood (oils derived from the bark) cluster with the abiotic samples, and even cyanobacteria seem similar in this view. Notice that the dried oak leaf, clearly a degraded living material, is remarkably similar to a cysteine (amino acid) and glucose Maillard reaction (used in the searing of foods to create flavors). A number of the natural materials that were classified as of biological origin or containing material of biological origin, also cluster closely with the abiotic samples, such as Quisqueite and Asphaltum. The peat sample (labeled natural) is placed between the bulk of both biological and natural samples.
Why should this technique work to classify samples according to the type labels? It has been argued that living things are both complex, but composed of molecules that occupy a relatively small space of possible diversity. [Work by Lee Cronin’s group has looked at the way biology restricts the possible structures of organic molecules to create complex macromolecules using few subunits. For example, the amino acid glycine is both important as a constituent of proteins, forming much of the structure of collagen, and is central to several biosynthesis pathways, that include the synthesis of porphyrins and thence to heme in red blood corpuscles. Some macromolecules such as cellulose are formed entirely of D-glucose, as are most complex sugar macromolecules. Cronin calls his technique Assembly Theory [1].]
But larger molecules constructed of a small number of simpler molecules alone are insufficient. Cellulose is a polymer of D-glucose molecules, but clearly, we would not state that a sheet of wet paper was once living, or formed by natural processes. A minimal complexity is required. Life relies on a suite of molecules connected by metabolic pathways that exquisitely restrict the possible number of resulting molecules, however complex, such as proteins that are constructed from just 20 of the possible much greater number of amino acids. At the heart of all life is the Krebs cycle which autotrophs use in the reverse direction to oxidation as part of carbon fixation to build biomass, often glucose to build cellulose cell walls.
The Pyr-GC-EI-MS technique detects a wide range of organic molecules, but the machine learning algorithm uses a set of specific ones to detect the requisite complexity as well as the abiotic randomness. In other words, this is complementary to Cronin’s “Assembly Theory” of life.
I would note that the PCA uses just 20 variables to separate the abiotic and biotic/natural samples. This appears adequate in the majority of the sample set but may be fewer than the variables used in the Random Forest machine learning algorithm. [A single Decision Tree using my reduced data uses just 12 rules – (masses and normalized frequency), but the accuracy is far lower. The Random Forest using different rules (masses and quantities, would be expected to use more features.]
How robust is this analysis?
The laboratory instrument generates a large amount of data for each sample, over 650 mass readings repeated over 6000 times over the scan time. The data was reduced for testing which in this case was 8149 values. There were 134 samples, 59 were classed as biotic or natural, and 75 were abiotic samples. A Random Forest (a suite of Decision Trees) algorithm proved the best method to classify the samples. This resulted in a 90+% correct classification of the sample types. The PCA visualization in Figure 2 is instructive as it shows how the samples were likely classified by the Random Forest model, and which samples were likely misclassified. The PCA used just 20 of the highest-scoring variables to separate the 2 classes of samples.
Generally, the Pyr-GC-EI-MS technique is considered robust with respect to masses extracted from different samples of the same material. The authors included replicates in the samples which should, ideally, be classified together in the same leaf in each Decision Tree in the Random Forest. That this is the case in this experiment is hinted by the few labels that point to 2 samples that are close together in the PCA shown in Figure 2, e.g. the cysteine-glucose Maillard reaction. That replicates are very similar is important as it indicates that the sample processing technique reliably produces the same output and therefore single samples are producing reliable mass and time signals with low noise. [In my experiment (see Appendix A) where K-means clustering was used, in most cases, the replicate pairs were collected together in the same cluster indicating that no special data treatment was needed to keep the replicates together.]
The pyrolysis of the samples transforms many of the compounds, often with more species than the original. For example, cellulose composed purely of D-Glucose will pyrolyze into several different compounds [18]. The assumption is that pyrolysis will preserve the differences between the biotic and abiotic samples, especially for material that has already undergone heating, such as coal. As the pyrolysis products in the mass range of 50 to 200 may no longer be the same as the original compounds, this technique can be applied to any sample containing organic material.
The robustness of the machine learning approach can be assessed by the distribution of the accuracy of the individual runs of the Random Forest. This is not indicated in the article. However, the high accuracy rate reported does suggest that the technique will report this level of accuracy consistently. What is not known is whether this existing trained model would continue to classify new samples accurately. This will also indicate the likely boundary conditions where this model works and whether retraining will be needed after the sample set is increased. This will be particularly important when assessing the nature of any confirmed extraterrestrial organic material that is materially different from that recovered from meteorites.
The robustness may be dependent on the labeling to train the Random Forest model. The sample set labels RNA and DNA as abiotic because they were sourced from a laboratory supply, while the lower complexity insect chitin exoskeleton was labeled biotic. But note that the chitin sample is within the abiotic bounding box in Figure 2, as well as the DNA sample.
Detecting life from samples that are fossils, degraded material, or parts of an organism like a skeletal structure, probably requires being able to look for both complexity and material that is composed of fewer, simpler subunits. In extremis, a sample with few organic molecules even after pyrolysis will likely not be complex enough to be identified as biotic (e.g. the meteorite samples), while a large range of organic molecules may be too varied and indicate abiotic production (e.g. Maillard reactions caused by heating). There will be intermediate cases, such as the chitinous exoskeleton of an insect that has relatively low molecular complexity but which the label defines as biotic.
What is important here is that while it might be instructive to know what the feature molecules are, and their likely pre-heated composition, the method does not rely on anything more than the mass and peak appearance time of the signal to classify the material.
Why does the Random Forest algorithm work well, and exceed that of a single Decision Tree or 2-layer Perceptron [a component of neural networks used in binary classification tasks]? A single Decision Tree requires that the set of features have a strong common overlap for all samples in the class. The greater the overlap, the fewer rules are needed. However, a single Decision Tree model is brittle in the face of noise. This is overcome with the Random Forest by using different subsets of the features to build each tree in the forest. With noisy data, this builds robustness as the predicted classification is based on a majority vote. (See Appendix A for a brief discussion on this.)
Is this technique agnostic?
Now let me address the important issue of whether this approach is agnostic to different biologies, as this is the crux of whether the experimental results will detect not just life, but extraterrestrial life. Will this approach address the possibly very different biologies of life evolved from a different biogenesis?
Astrobiology, a subject with no examples, is currently theoretical. There is almost an industry trying to provide tests for alien life. Perhaps the most famous example is the use of the disequilibria of atmospheric gases, proposed by James Lovelock. The idea is that life, especially autotrophs like plants on Earth, will create an imbalance in reactive gases such as oxygen and methane that keeps them apart from their equilibrium. This idea has since been bracketed with constraints and additional gases, but the basic idea remains a principal approach for exoplanets where only atmospheric gas spectra can be measured.
As life is hypothesized to require a complex set of molecules, yet far fewer than a random set of all possible molecules, or as Cronin has suggested, reuse of molecules to reduce the complexity of building large macromolecules, it is possible that there could be fossil life, either terrestrial or extraterrestrial, that has the same apparent complexity, but largely non-overlapping molecules. The Random Forest could therefore build some Decision Trees that could select different sets of molecules to make the same biotic classification, suggesting that this is an agnostic method. However, this has yet to be tested as there are no extraterrestrial biotic samples to test. It may require such samples, if found and characterized as biotic, to be added to a new training set should they not be classified as biotic using the current model.
As this experiment assumes that life is carbon-based, clearly truly exotic life based on other key elements such as silicon would be unlikely, but not impossible, to be detected if volatile non-organic materials in a sample could be classified correctly.
The authors explain what agnostic in their experiment means:
Our Proposed Biosignature is Agnostic. An important finding of this study is that abiotic, living, and taphonomic suites of organic molecules display well-defined clusters in their high-dimensional space, as illustrated in Fig. 2. At the same time, large “volumes” of this attribute space are unpopulated by either abiotic suites or terrestrial life. This topology suggests the possibility that an alien biochemistry might be recognized by forming its own attribute cluster in a different region of Fig. 2—a cluster that reflects the essential role in selection for function in biotic systems, albeit with potentially very different suites of functional molecules. Abiotic systems tend to cluster in a very narrow region of this phase space, which could in principle allow for easy identification of anomalous signals that are dissimilar to abiotic geochemical systems or known terrestrial life.
What they are stating is that their approach will detect the signs of life in both extant organisms and the resulting decay of their remains when fossilized, such as shales and fossil fuels like coal and oil. As the example PCA of Figure 2 shows, the abiotic samples are tightly clustered in a small space compared to the far greater space of the biotic and once-biotic samples. The authors’ Figure 1 shows that their chosen method results in fewer different molecules found in the Allende meteorite compared to a microbe. I note that the dried oak leaf that is also within the abiotic cluster of the PCA visualization is possibly there because the bulk of the material is cellulose. Cellulose is made of chains of polymerized D-glucose, and while the pyrolysis of cellulose is a physical process that creates a wider assortment of organic compounds [18], this still limits the possible pyrolysis products.
This analysis is complementary to Cronin’s Assembly Theory which theorizes a reduced molecular space of life compared to the randomness and greater complexity of purely chemical and physical processes. This is because life constrains its biochemistry to enzyme-mediated reaction pathways. Assembly Theory [1] and other complexity theories of life [15] would be expected to reduce the molecular space compared to the possible arrangements of all the atoms in an organism.
The authors’ method is probably detecting the greater space of molecules from the required complexity of life compared to the simpler samples and reactions that were labeled as abiotic.
For any extraterrestrial “carbon units” that are theorized to follow organizing principles, this method may well detect extraterrestrial life, whether extant or fossilized, from a unique abiogenesis. However, I would be cautious of this claim simply because there were no biotic extraterrestrial samples used, because we have none, only presumed abiotic samples such as the organic material inside meteorites that should not be contaminated with terrestrial life.
The authors suggest that an alien biology using very different biological molecules might form their own discrete cluster and therefore be detectable. In principle, this is true, but I am not sure that the Random Forest machine learning model would detect the attributes of this cluster without training examples to define the rules needed. Any such samples might simply expose any brittleness in the model and either cause an error or be classified as a false positive for either a biotic or abiotic sample. Ideally, as Asimov once stated, the phrase most associated with interesting discoveries “is not ‘Eureka’ but ‘That’s funny . . .’”, might be associated with an anomalous classification. This might be particularly noticeable if the technique indicates that the sample is abiotic, while a direct observation by microscope clearly shows wriggling microbes.
In summary, it is yet to be tested against new, unknown samples to confirm whether it is both robust, and also agnostic, for other carbon-based life.
The advantage of this technique for remote probes
While the instrument data would likely be sent to Earth regardless of local processing and any subsequent rover actions, the trained Random Forest model is computationally very lightweight and easy to run on the data. Inspection of the various Decision Trees in the Random Forest allows an explanation for which features best classify the samples. As the Random Forest is updated by larger sample sets, it is easy to update the model to analyze samples in the lab or on a remote robotic instrument, in contrast to Artificial Neural Network architectures (ANN) that are computationally intensive. Should a sample that looks like it could be alien life but produces an anomalous result (That’s funny…”), the data can be analyzed on Earth and then assigned a classification, and the Random Forest model rerun with the new data either on Earth and the model uploaded, or locally on the probe.
Let me stress again that the instrumentation needed is already available for life-detection missions on robotic probes. The most recent is the Mars Organic Molecule Analyzer (MOMA) [9] which is to be one of the suite of instruments on the Rosalind Franklin rover as part of the delayed ExoMars mission which is now planned for a 2028 launch. MOMA will use both the Pyr-GC-EI-MS sample processing approach, plus a UV laser on the organic material extracted from 2-meter subsurface drill cores to characterize the material. I would speculate that it might make sense to calibrate the sample set with the MOMA instruments to determine if the approach is as robust with this instrument as the lab equipment for this study. The sample set can be increased and run on the MOMA instruments and finalized well before the launch date.
[If the Morningstar Mission to Venus does detect organic material in the temperate Venusian clouds, perhaps in 2025, this type of analysis using instruments flown on a subsequent balloon mission might offer the fastest way to determine if that material is from a life form before any later sample return.]
While this is an exciting, innovative approach to classifying organic molecules and classifying them as biotic or abiotic, it is not the only approach and should be considered complementary. For example, terrestrial fossils may be completely mineralized, with their form indicating origin. A low-complexity fragment of an insect’s exoskeleton would have a form indicative of biotic origin. The dried oak leaf in the experiment that clusters with the abiotic samples would leave an impression in the sediment indicative of life, just as we see occasionally in coal seams. Impressions left by soft-bodied creatures that have completely decayed would not be detectable by this method even though their shape may be obviously from an organism. [Although note that shape alone was insufficient for determining the nature of the “fossils” in the Martian meteorite, ALH84001.]
Earlier, I mentioned that the cellulose of paper represents an example with low complexity compared to an organism. However, if a robot probe detected a fragment of paper buried in a Martian sediment, we would have little hesitation in identifying it as a technosignature. Similarly, a stone structure on Mars might have no organic material in its composition but clearly would be identified as an artifact built by intelligent beings.
Lastly, isotopic composition of elements can be indicative of origin when compared to the planetary background isotopic ratios. If we detected methane (CH4) with isotope ratios indicative of production by subsurface methanogens, that would be an important discovery, one that would be independent of this experimental approach.
Despite my caveats and cautions, local life detection, rather like the attempts with the 1976 Viking landers may be particularly important now that the Mars Sample Return mission costs are ballooning and may result in a cancelation, stymying the return to Earth of the samples Perseverance is collecting [16]. One of the major benefits of training the Apollo astronauts to understand the geology and identify important rock samples was the local decisions made by the astronauts over which rock samples to collect, rather than taking random samples and hoping the selection was informative. A mission to an icy moon would benefit from such local life detection efforts if multiple attempts need to be made in fairly rapid succession without requiring communication delays with Earth for analysis and decision-making and where no sample return to Earth was likely. This innovative technique appears to be an important contribution to the search for extraterrestrial life in our system, and possibly even beyond if our probes capture samples from interstellar objects.
The paper is “Cleaves, J et al, Hazen, R, “A robust, agnostic molecular biosignature based on machine Learning,” PNAS 120 (41) (September 25, 2023) e2307149120. Abstract.
———————————————————————
Appendix A. My experiment with the supplied data. [12]
Method
To test some of the feedback from the authors, I ran some simple machine-learning experiments on the data. Rather than reduce the data to the number of variables in the paper, I used a simple data reduction by collapsing the scan data dimension so that only the single mass values remained. I normalized to the largest mass value in a sample that was set to 100 and all normalized floating point numbers were reduced to integers. All the resulting values of less than 1 were therefore set to 0. I used the classification labels as given. I also shuffled the class labels to test that the information in the data was lost with this operation. I used the Weka ML software package for running Decision Trees, Random Forests, and other ML methods [20].
Results and Discussion
Using the example I ran [figure 3] it is clear that the presence of a molecule[s] of mass 280 is sufficient to classify 14 of the 59 biological samples with no other rules needed, and if that rule fails, passing a rule with the presence of a molecule about ½ the mass of the first rule, adds a further 8 samples correctly classified as biological. However, it takes a further 6 rules to classify another 22 biological samples, and 7 rules to select 48 (1 sample was a false positive) of the 75 abiotic samples. The rules used mostly used larger molecules to determine the classifications because they had the most discriminatory power, as suggested by the number of the larger molecules of the 20 used in the PCA visualization. Of the 12 rules in my experiment, all but 3 used masses of 100 or greater, with 3 rules of 200 or greater. It should be noted that many rules simply needed the presence or absence (less than 1% of the peak frequency) of a molecule. The 2 largest biotic and abiotic leaves each required 7 rules, but about half required some non-zero value. The biotic leaf with 22 samples had just 3 rules with peak values that were present, while the abiotic leaf with 49 classified samples had all 7 rules with no peak value or values below a threshold.
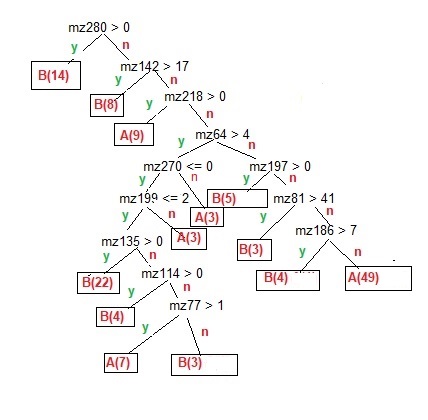
Figure 3. The model for a Decision Tree output for a reduced collapsed set of data. It shows the rule tree of different mass normalized frequencies to classify abiotic [A], and biotic and natural [B], samples as leaves. There are 134 samples, For training, all the samples were used, 75 are classed abiotic, and 59 and biotic/natural. [The few misclassified samples were excluded for simplicity and clarity]. As all samples were used, there was no out-of-sample testing of the model.
The best classifier was the Random Forest, as found by the authors. This far exceeded a single Decision Tree. It even exceeded a 2 layer Perceptron. The Random Forest managed to reach a little more than 80% correct classification, which fell to random with the shuffled data. While the results using the more greatly reduced data were less accurate than those of the paper, this is expected by the data reduction method.
To test whether the data had sufficient information to separate the 2 classes simply by clustering, I ran a K-Means clustering [14] to determine how the data separated.
1. The 2 clusters were each comprised of about 60% of one class. Therefore while the separation was poor, there was some separation using all the data. Shuffling the labels destroyed any information in the samples as it did with the Decision Tree and Random Forest tests.
2. The replicate pairs almost invariably stayed in the same cluster together, confirming the robustness of the data.
3. The natural samples, i.e. those with a biogenic origin, like coal, tended mostly to cluster with the abiogenic samples, rather than the biotic ones.
I would point out that the PCA in Figure 2 was interpreted to mean that abiotic samples clustered tightly together. However, an alternative interpretation is that the abiotic and natural samples separate from the biotic if a separation is drawn diagonally to separate the biotic samples from all the rest.
One labeling question I have was placing the commercially supplied DNA and RNA samples in the abiotic class. If we detected either as [degraded] samples on another world, we would almost certainly claim that we had detected life once the possibility of contamination was ruled out. Switching these labels made very little difference to my Random Forest classification overall, but it did switch more samples to be classified as biotic, in excess of the switch of the 2 samples to biotic labels. It did make a difference for a simpler Decision Tree. It increased the correct classifications (92 to 97 of 134), mostly reducing the misclassification of abiotic to biotic classes, (23 to 16). The cost of this improvement was 2 extra nodes and 1 leaf in the Decision Tree.
The poor results of the 2-layer Perceptron indicate that the nested rules used in the Decision Trees are needed to classify the data. Perceptrons are 2-layer artificial neural networks (ANNs) that have an input and output layer, but no hidden neural layers. Perceptons are known to fail the exclusive-OR test (XOR) although the example Decision Tree in Figure 3 does not require any variables to overcome this issue. A multilayer neural net with at least 1 hidden layer would be needed to match the results of the Random Forest.
In conclusion, my results show that even with a dimensionally reduced data set, the data contains some information in total that allows a weak separation of the 2 classification labels and that the random Forest is the best classifier of many that were available in the WEKA ML software package.
References
1. Assembly Theory (AT) – A New Approach to Detecting Extraterrestrial Life Unrecognizable by Present Technologies www.centauri-dreams.org/2023/05/16/assembly-theory-at-a-new-approach-to-detecting-extraterrestrial-life-unrecognizable-by-present-technologies/
2. Venus Life Finder: Scooping Big Science
www.centauri-dreams.org/2022/06/03/venus-life-finder-scooping-big-science/
3. Pyrolysis – Gas Chromatography – Mass Spectroscopy en.wikipedia.org/wiki/Pyrolysis%E2%80%93gas_chromatography%E2%80%93mass_spectrometry
4. Random Forest en.wikipedia.org/wiki/Random_forest accessed 10/05/2023/
5. PCA “Principal Component Analysis” en.wikipedia.org/wiki/Principal_component_analysis accessed 10/05/2023
6, SHERLOC “Scanning Habitable Environments with Raman and Luminescence for Organics and Chemicals“ en.wikipedia.org/wiki/Scanning_Habitable_Environments_with_Raman_and_Luminescence_for_Organics_and_Chemicals accessed 10/06/2023
7. Han, J et al, Organic Analysis on the Pueblito de Allende Meteorite Nature 222, 364–365 (1969). doi.org/10.1038/222364a0
8. Zenobi, R et al, Spatially Resolved Organic Analysis of the Allende Meteorite. Science, 24 Nov 1989 Vol 246, Issue 4933 pp. 1026-1029 doi.org/10.1126/science.246.4933.1026
9. Goesmann, F et al The Mars Organic Molecule Analyzer (MOMA) Instrument: Characterization of Organic Material in Martian Sediments. Astrobiology. 2017 Jul 1; 17(6-7): 655–685.
Published online 2017 Jul 1. doi: 10.1089/ast.2016.1551
10. Cleaves, J et al, Hazen, R, A robust, agnostic molecular biosignature based on machine Learning, PNAS September 25, 2023, 120 (41) e2307149120
doi.org/10.1073/pnas.2307149120
11. __ Supporting information. www.pnas.org/action/downloadSupplement?doi=10.1073%2Fpnas.2307149120&file=pnas.2307149120.sapp.pdf
12. __ Mass Spectroscopy data: osf.io/ubgwt
13. Gold, T. The Deep Hot Biosphere: The Myth of Fossil Fuels. Springer Science and Business Media, 2001.
14. K-means clustering en.wikipedia.org/wiki/K-means_clustering
15. Chou, L et al Planetary Mass Spectrometry for Agnostic Life Detection in the Solar System Front. Astron. Space Sci., 07 October 2021 Sec. Astrobiology Volume 8 – 2021
doi.org/10.3389/fspas.2021.755100
16. “Nasa’s hunt for signs of life on Mars divides experts as mission costs rocket“ Web access 11/13/2023 www.theguardian.com/science/2023/nov/12/experts-split-over-nasa-mission-to-mars-costs-rocket
17. The Astrobiology Field Laboratory. September 26, 2006. Final report of the MEPAG Astrobiology Field Laboratory Science Steering Group (AFL-SSG). Web: mepag.jpl.nasa.gov/reports/AFL_SSG_WHITE_PAPER_v3.doc
18. Wang, Q., Song, H., Pan, S. et al. Initial pyrolysis mechanism and product formation of cellulose: An Experimental and Density functional theory(DFT) study. Sci Rep 10, 3626 (2020). https://doi.org/10.1038/s41598-020-60095-2
19. Sharma, S., Roppel, R.D., Murphy, A.E. et al. Diverse organic-mineral associations in Jezero crater, Mars. Nature 619, 724–732 (2023). https://doi.org/10.1038/s41586-023-06143-z
20. Weka 3: Machine Learning Software in Java https://www.cs.waikato.ac.nz/ml/weka/



“Alien life or Chemistry ?” That is a good title.
As A. Tolley puts it “While we “know life when we see it”, nevertheless we still struggle to define what life is ..”
That’s the point.
The PNAS paper is completely wrong from an epistemological point of view.
They are back to the XIXth century reductionism.
They forget an essential instrumentation: natural language.
For natural language, “life” is a word with its cohort of emotions.
You cannot escape the empirical fact that scientists speak two languages:
natural language, with its emotional resonances, and mathematics.
As a consequence, there is no objective definition of “life”.
One cannot escape that the statement “this is living” is an arbitrary choice, or judgement,
similar to the judgement “this embryo is a human”.
My purpose is not to add to the numerous philosophical considerations,
it is socio-political: be prepared to strong battles, when in the next decades
we will have an accumulation of candidate “biosignatures”, battles between
“pro-life” and “pro-chemistry”.
Rendez-vous in 15-20 years.
Increasing the confidence level would require analysis of multiple samples. Too few and the results are going to be too ambiguous. However, there will be other cues, such as from cameras to visually analyze the samples. I also believe that analysis of samples drilled from rock will be very different from scoops of regolith or dirty ice.
Ambient conditions will affect the analysis, whether it be a surface or sub-surface sample, the atmosphere, volatiles in the ground and so forth.
It could prove difficult to get a high confidence result unless the sample jumps out of the scoop!
If only! I will settle for wriggling, waving cilia, a metabolizing and reproducing sample of cellular life, even a homochiral protein or two.
I do think they will need to extend the sample set to cover a wider range of materials, especially those that might sit on the edge between biotic and non-biotic origins. For example, material that is of biotic origin but is “simple”, like silk. Then there is the issue of the samples chosen in the experiment and the labeling. Should cedar oil be labeled as biotic, and biological macromolecules like DNA, and RNA, as abiotic because they were made artificially? Paper is relatively simple – mostly cellulose and lignin, and one would expect it would be similar in composition to tree bark, dried leaf, and even wood. Another issue is the degradation of biological material over time. Their natural materials such as petroleum products are altered by geological and biological processes, including heat.
[Back when I was an undergraduate, and mammoths still wandered in Siberia, a textbook questioned the source of oil. The evidence was equivocal. We have since settled on biological origin, but Thomas Gold’s “Deep Hot Biosphere” hypothesis suggested oil and coal was formed by geology and that the crustal biosphere contaminated and contributed to their composition. Had this experiment been down back then, how would all these samples be labeled? Lastly, we assume asteroidal amino acids are abiotic based on the knowledge that these AAs can be formed abiotically and that meteorite AA samples are not homochiral. But AAs can become racemic mixtures, especially over millions/billions of years.
As I say in the article, this looks like it could be a useful tool in the toolbox. However, it would be useless if faced with a marble statue, the Antikythera mechanism, and probably fossil ammonites, all of which could be identified as technosignatures using a simple camera and shape recognition software.
Even with abundant material, it can be very difficult to identify a sample. The red rain in Kerala is a reasonably contemporary example. If there are samples of this material available, it would be interesting to see how this technology would classify it.
. Given the range of samples and their conditions, I don’t think that would be an issue. My concern would be that the in situ processing of a sample, e.g. on Mars, would be different from that of the calibrated analysis on Earth. I am not sure how that would be handled, other than redoing the processing of the sample set where the new, unknown samples are taken. This may only be resolved by experiments, such as running the same sample both remotely and back on Earth.
I am going to have to disagree with you on this point. The edge cases are viruses, degraded fossil material, and technological artifacts (clothing, tools, ornaments) made from biological materials such as wood.
The question is how far samples differ from terrestrial biology will this technique work? Alien building blocks of life may be completely unidentifiable. Depending on the model captured by the decision trees in the Random Forest, an unidentifiable sample could fall into either bucket. Ideally one would want another bucket[s] to capture samples that would fall into one of the biotic/abiotic leaves of the tree[s] by default.
Where this technique could excel is making rapid determinations of probable origin as samples are taken from sites. Samples passing the biotic classification would then be subjected to scrutiny by other means, including possible sample return to Earth where possible. A lander on Ceres could take samples from areas of upwelling and those indicating possible biotic origin could be stored and returned to Earth.
Angsting over epistemology isn’t productive IMO. Perhaps you would care to flesh out what you would see as the problem in using this approach and whether your argument would invalidate previous searches.
From going to astrobiology conferences, and talking to biologists I came to realize that there is a big gulf between physicists and biologists about life. (And, of course, it ain’t going to be accepted as life until the biologists say it is.)
Physicists tend to think in terms like “if the concentrations of molecules X, Y and Z are > the background by N sigma” then LIFE !
In my experience, that does not really impress biologists, or at least, the biologists I talked to. They want to understand how the ecosystem works (e.g., where does free energy come from, how is it consumed, what are the cycles of nutrients, etc.). They also really want microscopes.
I’m a biologist and on the same page as those you’ve talked to.
The excess of any chemical is not indicative of life.
It show the presence of a chemical reaction, either powered by heat, natural catalysation, serpentinisation (which caused quite some stir due to detecting methane on Mars) and lastly enzymes.
But neither is alive in any sensible semantic use of the word.
This while some or several of those processes might have been involved in how life got started in the first place.
@Andrei
Have you looked at Cronin’s work, as well as others on related ideas, concerning the imbalance of specific compounds compared to their likely abundances based on chemistry. It is very much related to the idea of metabolic pathways and their possibly origination by autocatalytic sets. Essentially is is a form of chemical Darwinism.
When we did organic chemistry experiments in labs, yields of specific compounds were often low compared to a host of other compounds also created by the reaction process. But as we know from biology, enzymatic reactions are very specific and prevent almost all of the side reactions. That is the basis for the idea of the narrowness of the chemical space vs the chemical space due to chemistry.
Cronin’s “Assembly Theory” work sits on top of this by indicating that because of the biological pathways, larger molecules are built from the existing simpler ones, even if that is not the way chemists might try to construct those larger compounds. Again, just as evolution works with prior structures, resulting in somewhat klugey organisms, so we see this down at the biochemistry level inside cells.
My bottom line, is that this idea makes a lot of sense, even if I would still like to “see organisms with my own eyes” under the microscope. It is not so different from accepting genome analysis to piece together evolution, rather than using fossils. Information theory vs visual observation of forms.
A search for “life” based on looking at chemistry is quite valid, just as one based on looking for radio transmissions. The chemistry spans the range from complex organic to unequivocally biotic, and includes durable products, by-products, wastes and detritus. And products such as Dyson swarms would be stepping beyond chemistry.
When zooming in at the level of molecular cell biology, labelling an assemblage of molecules as “living” or otherwise may be problematic, but where one sets the border depends on one’s familiarity with the territory.
I resemble that remark. ;-)
I would say, yes and no. A newly expired organism is indistinguishable from a live one, unless the dynamics can be detected. Medics use EKGs and EEGs to make that determination. Once upon a time, Vitalists, thought that dead organisms could be revived by extrapolating Galvani’s electrical experiments – hence Frankenstein’s monster.
Evolutionists don’t care about dynamics, just the form of dead organisms, and in the last couple of decades, the information in the genomes of living organisms (and from the “recently” preserved.)
Measurements can be very useful, but one must be wary. A few years ago there was the rather embarrassing case of fMRIs from a dead salmon. [Forttunately that was fixed with an algorithm change. But it should be a lesson on relying on models.]
For searches of organisms that should be extant, rather than extinct- e.g. in the subsurface oceans of icy worlds, then I think a microscope would be useful. Digital microscopes of various types can be very small and low mass. Why not add one to the instrument suite? If we redid the Viking experiments a microscope would have resolved the ambiguity pretty quickly, especially if there wriggling bugs to be seen. Unfortunately one really needs a [transmission] electron microscope to see the details of bugs and the components of eukaryotic cells. This might be the case if the cells in Enceladan plumes have been broken open. In this case, a sample return would be needed.
But sometimes proxies are useful. Keeling’s CO2 measurements on Hawaii show the annual changes due to the axial tilt of the Earth, the imbalance of geography with the predominance of northern hemisphere temperate and boreal forests. That simple measurement might be ambiguous for an exoplanet, but it would be worth following up with other proxy measurements.
The Cleaves and Hazen paper is more interesting to me in that it allows local analysis of organic material, potentially increasing the rate of sample testing and local decision-making by a rover. You may recall the field training of the Apollo 15 astronauts that allowed them to collect the most interesting rocks on the Moon. (c.f. “From the Earth to the Moon” episode 10 “Galileo Was Right” for a dramatization). This is what one wants from a good “field lab rover/probe” – putting real domain expertise into a machine. It is the heart of the argument between astronauts vs robots in space exploration.
But no measurement is definitive. Biologists argue over whether viruses are alive or not. There is no, as yet, means to answer that question. [Although, unlike Jean Schneider’s comment above, I don’t think it is that important. Viruses wouldn’t exist without life, and therefore could be considered probes for the presence of life.] But a wriggling bug, or a growing, replicating colony of bugs, clearly seen in a microscope…
@Robin
One issue is the life/not-life point. I consider it a gray area, like “intelligence” whether organismal or artificial. Is abiogenesis a “phase change” that occurs rapidly, or a slow creative process? If metabolism is the key, then Stuart Kauffman’s work on autocatalytic sets suggests something more akin to a phase change once enough molecules participate. This might be similar to the RNA-World scenario too, although the creation of RNA and its analogs is very much more difficult. And all this has less to do with biology than other disciplines.
At any transition point, any purely simple molecular analysis should not be able to distinguish between chemistry and life.
We’re going to have constant argument once we get samples to analyze as we should. This is one of the holy grails of biology. Define life as exactly as possible and find it elsewhere (or strong evidence it existed previously). Surely the most defining feature of life is its ability to reproduce or had the ability to reproduce when alive. The rest of the requirements such as having a metabolism are a lot more difficult to absolutely prove if the samples are no longer alive. I continue to be fascinated by the ALH84001 case. Is there a final scientific consensus on this? I like the inclusion of edge samples but I think we’re after things that are solidly “alive”, in particular something similar to our prokaryotes or eukaryotes. Whether we recognize things that are well beyond our own experience of nature on Earth is hard to say. A chemical approach such as this can only help at least initially in establishing boundaries.
@Gary
You can read the Wikipedia entry Allan Hills 84001 on its status. TL;DR is that the morphology was insufficient to prove the forms were fossil life, but there are still adherents to that interpretation. We have a similar issue with some terrestrial “fossils” – “are they or aren’t they?”.
Thank you Alex. A final word from me about ALH84001. There is no scientific consensus as far as I can tell about this sample. However the strongest likelihood from the analyses done so far is that the rock does not contain the remnants of previously living organisms. This may be the type of sample in or on which we will have to continue to try to determine whether living or formerly living material is present. It will continue to be studied I’m sure as further analysis has revealed further interesting qualities. Having an age of about 4 billion years and being from part of the Valles Marineris from about 17 million years ago and likely in shallow water before being sent away from Mars by a likely meteorite strike make this sample extremely valuable and extremely rare.
As someone who has worked with PCA and RF in a Remote Sensing context, I have to say the following: Principal component Analysis is great at finding separability IN THE ORIGINAL DATA. I will give N axes, where N is the number of bands in the image, of separability. The first axis will have something like 70% of what makes the samples unique, by the third it is over 95% typically and 6 axes are necessary only if we are talking about hundreds of bands. The problem then becomes what is exactly that this image stack is showing you in the PCA axes. If we are talking about a vegetated landscape, PCA can often mean just a fancier way to show what NDVI or EVI will show you. Also the problem is that when you enter another image stack, the PCA axis is not going to be the same and it could be down to sensor issues or even just noise in the image, not necessarily something that shows actual difference on the ground.
The best thing about Random Forests is the cross validation. I am not sure if that option is available in your package, but on what we used at grad school, we would turn it on and then selection that option when the trees were out. Cross validation would prune the tree to 4 or 5 branches rather than the 20-30 branch tree we had originally. RF had the advantage that these trees could then be used with new data so long we had the same sensor that was similarly calibrated and corrected for atmospheric and geometric effects. That was not the case with PCA
Honestly you could put the alien sample and try to classify along with biotic and abiotic samples from earth to see where it is closer. The best you would still get is something like a Bayesian percentage based on what the other samples were that you chose. Closer to this or to that axis and then you should expect to endure papers from other authors who would create their own biotic to abiotic axis from different materials to see where it is classified for them. I doubt you would get a clear cut answer but then again, that is how science works anyway
@Ioannis
I had not come across PCA until I worked in biotech and the scientists loved using 3-axis PCA to show the effect of chemicals on organisms based on gene expression. Visualization is a very powerful means to show similarities and differences. Other methods include dendograms (but I found that the scientists had a poor understanding of what they showed, even though this is a very common 2-D representation), and various clustering methods (e.g. K-means that I used in the Appendix) but simple K-means require selecting the number of clusters, whilst PCA effectively avoids that issue. Who doesn’t like to “press a button” and get an answer?
The use of 2D PCA in the journal paper is for a visual representation to show the separation of abiotic from biotic samples. It also shows how some biotic labeled samples seem to be in the abiotic cluster and warrants understanding why.
Idk what the Matlab code used in teh paper does with reporting Random Forest output, but the package I use just gives the binary interpretation. It would be nicer if it could give probability estimates for each sample, as some ANN packages can offer. The problem with RFs is that one cannot know how the validation was done, nor if boosting was used, which can affect the probabilities. The advantage is that it, like individual decision trees, can be computed relatively quickly as it uses a greedy algorithm to separate the data. Theoretically one could inspect each individual tree output to try to understand the output of the forest. In my experience, single decision trees are preferred, even if the output is more brittle, and may give a false impression of the features used to separate the samples.
As for getting an extraterrestrial sample to test, we are still waiting. Ideally, we want an alien sample that we know is biotic through some other means (camera, microscope) and test that. Is it classified as biotic or not?
What a great study, citizen data scientist Tolley!
Training the model on Mars and Enceledus where we can confirm or refute the results by returning samples to earth for follow-up analysis should improve the accuracy for analysis of data for exoplanets. Can the ML analysis of chemistry-set data from our solar system then be applied to spectral analysis of light from distant planets?
Since I had mentioned Kauffman and autocatalytic sets, this Arxiv paper has emerged:
Is the Emergence of Life an Expected Phase Transition in the Evolving Universe?
This paper marries 2 concepts to suggest the mechanism for abiogenesis, as well as a test for life.
It is a pity that the Cleaves and Hazen paper uses pyrolysis as this can alter the source molecules. If purely chromatography and mass spec could be used, then Kauffman’s suggested experiment is possible (although I think other methods may be needed to characterize some molecules.)
Not mentioned in the Kauffman paper, but worth checking experimentally, is whether chiral molecules (or a subset) that participate in autocatalytic sets need to be homochiral or not. The molecular graph in Kauffman’s Figure 3 could be evaluated for the chiral molecules to test this hypothesis.
[Kauffman also notes another recent PNAS paper by Cleaves and Hazen on the evolution of functionality. ]
What an intriguing paper. The early appearance of life on Earth may be explained by the broad array of geophysical processes present. The diversity of chemicals and reactions would have been immense. This approach could also explain the jump in complexity following global ice events. The interface between ice, water and land would be a massive diversity driving factory of chemicals and catalyzing pathways.
The assumption that the evolution of a biome is fundamentally unpredictable may also provide a motivation for non-interference. All we need to do is assume that a biome produces valued assets, for instance qualia that an ETI could experience in virtual reality or add to themselves in base reality. The probability that a similar supply could be found elsewhere would be vanishingly low.
The combination of unique starting conditions, unpredictable evolution, and path dependency also raises the possibility that a biome in base reality could be used to seed biomes in virtual reality. A possibility I am studiously resisting.
@Harold.
There are 2 interesting issues that fascinate me in the evolution of complex life:
1. Why were all the major phyla created in the “Cambrian explosion”? What allowed these very different body plans to be developed and how long did it take? The prior Ediacaran fossils do show this variety, suggesting that it was a relatively rapid process. (I don’t think it was “aliens” ;P )
2. The emergence of metamorphosis in many genera. There are a number of theories, but while intriguing, need more work to establish which mechanisms operate. The recent popular books on the subject The Mystery of Metamorphosis (2011) and Larvae and Evolution (1992) are both very old school biology, with no genome and gene expression analysis to support the ideas. [To be fair. L&E is pre-genomic analysis era in biology. TMoM cannot make that excuse.]
Interesting follow-up on my analysis. I wanted to keep the value of the scan number as well as the masses. I changed the maximum normalized value from 100 to 1000, so that I could still discard values of less than 1% of the maximum (i.e. 10).
To reduce the size of the feature set, I used 2 criteria:
1. reduced the number of variables by calculating the Gini value for each feature across the data. I filtered for low values, i.e. furthest from random.
2. Because my normalization created many features (scan-mass) with 0 values, I used a variable for the number of samples that would make the cut for each variable when calculating the best split for the data.
Results:
1. Disappointingly, the use of the Gini value to filter for good variables did not improve the scores for the decision tree or Random Forest ML classification.
2. The features selected by the algorithm was highly dependent on the number of samples that were included in the “selected for” split. This selected for lower mass values. By reducing the constraint on number of samples in the “selected for” split, the larger the mass sizes used for the ML, although this quickly reached a maximum well below the masses selected by the journal article.
The last constraint indicated that the samples had a relatively low overlap when the data was reduced by filtering out values below 1% of the maximum. IOW, for any scan-mass variable, most of the samples would have their values set to 0. The lower masses, around 50-70 were those that most easily passed tight constraints on numbers.
My conclusion is that eliminating values that might be considered in the noise regime, resulted in the samples having poor overlap for values between 1-99% of the maximum. If this is valid, it might argue for the high probability of correct classification being based on very low values which may not be reproducible. [It is also possible that my approach to data reduction creates artifacts, and also that my code may be flawed when building the data file for the WEKA ML package that I used.]
I will investigate further as time permits. If the data reduction by removing the values <1% of the maximum is an artifact, I can relax this constraint to 0.1%, or less. The decision tree rules using these low-value variables should be evident.
What I want to do is explore whether the Assembly Theory concept can be extracted from the data. The idea is that this theory should indicate fewer peaks for each mass as the scan progresses, i.e. that the number of different compounds that emerge by the gas chromatography should be lower per mass bin than random chemistry. Obviously, this is somewhat corrupted by the pyrolysis process, but just maybe sufficient integrity of subunits for each macromolecule. The data set clearly contains masses in the 300-unit range, which if pure carbon would indicate molecules with ~25 carbon atoms. If they are composed of relatively few units of lower mass, this would suggest the Assembly Theory for the process life uses may be detectable, compared to non-living, chemical/physical processes.
Experiment update.
Despite playing with the normalization and filtering settings, I could not get any higher correct classification above very low 80’s%. This is despite using both the mass and scan# to uniquely identify the variables (in the millions). The Gini value to reduce the data was not effective other than to reduce the number of variables and hence memory space. So sadly I was not able to replicate the authors’ analysis.
I did run my proxy for Assembly Theory by:
1. Using the normalized data filtered by Gini values as before.
2. Maintaining a list of masses that had any non-zero values throughout all the scans.
3. Saving the binary list of masses, and the number of hits for each mass through the scans, and calculating overall numbers of masses, features (mass & scan value > 0), and the feature/mass ratio.
The idea is to ignore the normalized abundances and just assume that each hit for the mass in a scan represents a different compound emerging from the GC. Theoretically, the ratio for the number of hits for each mass should be lower for biotic than abiotic material.
I used the below 1% normalized mass as the minimum acceptable presence of the compound. I also filtered the mass range to accept up to 300 units. I did not filter the scan # range.
Results
1. The decision tree reached 77% correct, and the RF 80% correct – broadly the same as using the normalized data.
2. The decision tree used more rules than the original normalized abundances approach.
3. Both the presence of a mass in the data, and the number of hits per mass variables were both used in the tree.
4. The highest informative rule used the number of hits (>9) for a high mass (280) to classify 22 samples as biotic. More samples were classified as biotic with hits greater than those for abiotic samples. A mass of 99 had a lot of hits and was the last rule in a tree that created the 2nd largest leaf of 12 samples classified as biotic if the hits were <= 1006.
5. Almost all the rules used the presence of a mass and hit ratios for a mass where the mass was mostly in the 200-300 range, with the notable exception of mass = 99. {see output at the end.)
While it appears that the hits ratio falsifies the Assembly theory, it is possible that what this shows is that for any given mass, it is possible that there are strong peaks that result in many hits for the same compound across scans, rather than indicative of many compounds, each unique for each scan.
Conclusion
The presence of a mass in the spectrum (not its abundance) and the number of hits for that mass across all the scans contain the discriminatory data needed to correctly classify the samples with an accuracy comparable to using the normalized masses for each scan.
Decison Tree output:
[mz# = binary presence of the mass# with a non-zero value after normalization and removal of all values <1% of the maximum normalized value.
f_# = number of hits for the mass#. This is the number of non-zero values through all the scans for that mass value.]
f_280 <= 9
| mz192 0
| | mz194 0
| | | mz128 0
| | | | mz251 <= 0
| | | | | f_248 <= 13
| | | | | | mz266 <= 0
| | | | | | | mz290 <= 0
| | | | | | | | mz269 <= 0
| | | | | | | | | f_265 <= 1
| | | | | | | | | | mz249 <= 0
| | | | | | | | | | | mz244 <= 0
| | | | | | | | | | | | f_99 1006
| | | | | | | | | | | | | mz286 0: B (3.0)
| | | | | | | | | | | mz244 > 0: A (4.0)
| | | | | | | | | | mz249 > 0: B (5.0)
| | | | | | | | | f_265 > 1: A (2.0)
| | | | | | | | mz269 > 0: B (4.0)
| | | | | | | mz290 > 0: A (2.0)
| | | | | | mz266 > 0: B (7.0)
| | | | | f_248 > 13: A (4.0)
| | | | mz251 > 0
| | | | | f_148 <= 33
| | | | | | f_165 22: B (5.0)
| | | | | f_148 > 33: A (24.0/1.0)
f_280 > 9: B (22.0)
Number of Leaves : 17
Size of the tree : 33
=== Stratified cross-validation ===
=== Summary ===
Correctly Classified Instances 103 76.8657 %
Incorrectly Classified Instances 31 23.1343 %
=== Detailed Accuracy By Class ===
TP Rate FP Rate Precision Recall F-Measure MCC ROC Area PRC Area Class
0.787 0.254 0.797 0.787 0.792 0.532 0.796 0.770 A
0.746 0.213 0.733 0.746 0.739 0.532 0.796 0.732 B
Weighted Avg. 0.769 0.236 0.769 0.769 0.769 0.532 0.796 0.753
=== Confusion Matrix ===
a b <– classified as
59 16 | a = A
15 44 | b = B
Testing the Assembly Theory using the Cleaves and Hazen data.
The idea is to flag as a hit if a particular mass is recovered and the number of times the scan finds a value for that mass. Theoretically, biotic samples should have lower numbers of hits per mass than abiotic samples.
Method: normalize an filter data as before, but extract profiles through the scans for a particular mass.
[Note that I found that with less than 1% of maximum values discarded, there was still a residual 2-3% of noise. Raising the filter to 5% reduced the data greatly as ell as separating the peaks.
For the 6 samples tested – glycine (mol wt 76). glycine with Maillard Reaction, DNA, RNA, Pittston Coal, and cyanobacteria, using the mol wt of 99 (mz99), each showed string, sharp peaks in the data. This indicated that the molecules were likely different from the base sample as they would be isolated by the chromatography process. Sharp peaks indicate distinct discrete, compounds.
Some findings [Sorry I cannot show the chart images].
1. For the few samples tested, there did seem to be an indication that samples that one expects to be biotic showed fewer peaks – i.e. numbers if discrete peaks – for the 99 masses. Much more data needs to be extracted to confirm this, but see the decision tree below.
2. Glycine that has a mol wt of 76 should not appear at all, yet it does, with one sharp peal around scan 1200, and a “hump” around scan 2200 indicating a lot of very similar compounds? That the data is above its mol wt, this implies that glycine is forming new compounds with higher molecular wts.
3. Decision tree.
Most of the biotic samples 44 out of 59 have:
a mz60 gt 0, hits at mz55 gt 108 (ie greater multiple hits), hits mz281 lteq 1 (i.e. no multiple hits), hits at mz55 lteq 39 (ie fewer hits) and hits at mz87 lteq 28.
30 out of 75 abiotic samples mz60 gt 0, hits at mz55 lteq 108, mz178 lteq 0(i.e. missing), and mz64 lteq 0 (ie missing). A further 15 have mz60 lteq 0 (ie missing). The branch where mz160 gt 0 contains 20 correctly classified abiotic samples. (all the hits and greater at the selected masses than for the biotic samples.)
Overall, this suggests that the AT theory where larger biotic molecules are made of fewer discrete subunits than abiotic ones may be correct and captured by this technique. However, at this point, the peaks are not yet correctly reduced to single values, and am I not sure that the glycine hump is anything more than a pyrolysis artifact. [Any expert help would be appreciated.] Pyrolysis as a process may be confounding this data with noise due to reactions that we know happen when heating molecules that can facilitate reactions.
[Figure 1 in the paper and shown in the post has a mass cutoff well below 700, and the scans are much less than the 6400+, in the data. I noted the mass cutoff in my normalized data. The sample tests through the scans for the mass of 99 also show a rapid decline in peaks after around scan 3500. Why this truncating of the data was done was n]ot a question I asked of the team, but it emerged from working with the data.]
I do commend the authors for publishing their data as it permits independent analyses that may offer more insight into understanding the biotic vs abiotic classification question.
mz60 lteq 0: A (15.0)
mz60 gt 0
| mz160 lteq 0
| | f_55 lteq 108
| | | mz178 lteq 0
| | | | mz64 lteq 0: A (30.0)
| | | | mz64 gt 0
| | | | | mz68 lteq 0
| | | | | | mz94 lteq 0
| | | | | | | mz76 lteq 0: B (4.0/1.0)
| | | | | | | mz76 gt 0: A (7.0)
| | | | | | mz94 gt 0: B (7.0)
| | | | | mz68 gt 0: A (3.0)
| | | mz178 gt 0: B (3.0)
| | f_55 gt 108
| | | f_281 lteq 1
| | | | f_50 lteq 39
| | | | | f_87 lteq 28: B (44.0)
| | | | | f_87 gt 28: A (3.0/1.0)
| | | | f_50 gt 39: A (5.0/1.0)
| | | f_281 gt 1: A (4.0)
| mz160 gt 0: A (9.0)
lteq = less than or equal to
gt = greater than
Correctly Classified Instances 107 79.8507 %
Incorrectly Classified Instances 27 20.1493 %
=== Confusion Matrix ===
a b <– classified as
60 15 | a = A
12 47 | b = B
Possibility of lithopanspermy makes things much more complicated in the Solar system. There almost certainly was enough material exchange between surfaces of Venus, Earth and Mars, in the times when latter two and possibly the first one had readily habitable conditions on their surfaces. Identifying something living on Mars and in the clouds of Venus is comparatively easy, but figuring out what came from where, and how long ago, is a whole another level. Possibly only advanced molecular biology will tell when lines diverged and where it all started. Imagine we have remotely acquired data on Martian and Venusian microbes and know they, too, are made of proteins, sugars and nucleic acids. They differ from us too much to be surely from the same lineage, maybe even some different amino and nucleic acids and a number of unique and very unusual small molecules, signal, regulatory, etc. But no real “alien biochemistry”. Are these differences due to early divergence followed by 3 Gyrs of isolation and adaptation to extreme conditions? There could be some generalized metric of dissimilarity, akin to molecular clocks. What should be the difference of separate abiogenesis? How does it compare to the limits of divergence?
Imho, we could be sure enough about separate abiogenesis only after finding unequivocally complex disequilibrium biosignatures on exoplanets, or if we are lucky enough with something surely different on ocean floors of icy moons. Not sure which task is easier.
torque_xtr
At this point, we would be happy just to answer the question of whether a sample of organic material was from life or simply chemistry. [IMO, if you have any DNA/RNA, protein, or structured macromolecule, it is almost certainly from life, rather than chemistry. – SNA, RNA, peptides of some length, etc. are extremely hard to create even with directed chemistry with controlled conditions. However, what is the survival time of such macromolecules?]
The issue of the origin of life is going to be very interesting if we have samples that we can clearly identify as life, especially extant life, e.g. from a Martian cave, or even from an icy moon subsurface ocean. If the genetic code is the same for the samples – arrived at by synthesizing the DNA or RNA and translating the sequence into a functional protein, then I would venture that this suggests a common origin. (it may not if it turns out the code is an optimal one based on evolution, rather than a “frozen accident”). If we have living cells, so that the DNA can be fully sequenced, then using a subunit of ribosomal RNA (highly conserved) we can make estimates of when the lineages diverged. Without that, it will be hard to estimate divergence.
So
1. if we only have a sample of organic matter, we may be able to determine if the sample is abiotic or biotic.
2. If we have recognizable proteins, we may be able to make a guess at more details of the biotic source.
3. If we have DNA, we should be able to make far more inferences, including whether the lineages are separate or not, and if from a common ancestor, when divergence may have occurred.
4. If we have living cells (or ones that can be created from DNA pieces in the sample (very Jurassic Park!) we can do a lot more to understand the alien life collected.
But I stress, this all depends on whether we can get any samples at all, and how they can be analyzed. So far we have zip in terms of material that could be from a biotic source.
re: Experiment update:
Tree display error due to the interpretation of markup symbol “>”
For the f_99 hits it should be:
f_99 less than/equal to 1006 : B (12.0)
f_99 greater than 1006
| mz286 less than/equal to 0: A (6.0)
| mz286 greater than 0: B (3.0)